-Search query
-Search result
Showing 1 - 50 of 525 items for (author: liu & gb)

EMDB-34992: 
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens (UltrAuFoil)
Method: single particle / : Xiao J, Wang L
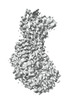
EMDB-39353: 
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens
Method: single particle / : Xiao J, Wang L

PDB-8hsb: 
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens (UltrAuFoil)
Method: single particle / : Xiao J, Wang L

PDB-8yjy: 
Cryo-EM Structure of CdnG-E2 complex from Serratia marcescens
Method: single particle / : Xiao J, Wang L

EMDB-35832: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

PDB-8iyx: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

EMDB-37795: 
Cry-EM structure of cannabinoid receptor-arrestin 2 complex
Method: single particle / : Wang YX, Wang T, Wu LJ, Hua T, Liu ZJ

PDB-8wrz: 
Cry-EM structure of cannabinoid receptor-arrestin 2 complex
Method: single particle / : Wang YX, Wang T, Wu LJ, Hua T, Liu ZJ

EMDB-16933: 
Structure of cGAS in complex with SPSB3-ELOBC
Method: single particle / : Xu PB, Ablasser A

EMDB-16936: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)
Method: single particle / : Xu PB, Ablasser A

EMDB-16937: 
Consensus refinement of cGAS/spsb3/EloBC/Nucleosome
Method: single particle / : Xu PB, Ablasser A

EMDB-16938: 
human cGAS/spsb3/EloBC in complex with nucleosome (2:2)
Method: single particle / : Xu PB, Ablasser A

PDB-8okx: 
Structure of cGAS in complex with SPSB3-ELOBC
Method: single particle / : Xu PB, Ablasser A

PDB-8ol1: 
cGAS-Nucleosome in complex with SPSB3-ELOBC (composite structure)
Method: single particle / : Xu PB, Ablasser A

EMDB-33990: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N
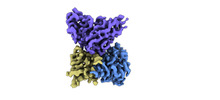
EMDB-33992: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

EMDB-33993: 
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Method: single particle / : Liu L, Sun H, Jiang Y, Liu X, Zhao D, Zheng Q, Li S, Chen Y, Xia N
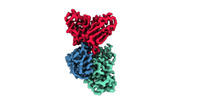
EMDB-33994: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yoy: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yp1: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yp2: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

EMDB-41856: 
Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A
Method: single particle / : Liu B, Kompaniiets D, Wang D

PDB-8u3b: 
Cryo-EM structure of E. coli NarL-transcription activation complex at 3.2A
Method: single particle / : Liu B, Kompaniiets D, Wang D
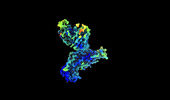
EMDB-35788: 
Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement)
Method: single particle / : Sun H, Jiang Y, Zheng Z, Zheng Q, Li S

EMDB-35789: 
Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (state 2)
Method: single particle / : Sun H, Jiang Y, Zheng Z, Zheng Q, Li S

EMDB-35966: 
Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (state 1)
Method: single particle / : Sun H, Jiang Y, Zheng Z, Zheng Q, Li S

PDB-8ix3: 
Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement)
Method: single particle / : Sun H, Jiang Y, Zheng Z, Zheng Q, Li S
EMDB-29645: 
Cryo-EM structure of an orphan GPCR-Gi protein signaling complex
Method: single particle / : Zhang X, Wang YJ, Li X, Liu GB, Gong WM, Zhang C

EMDB-40270: 
Cryo-EM structure of GPR34-Gi complex
Method: single particle / : Yong XH, Zhao C, Yan W, Shao ZH

PDB-8sai: 
Cryo-EM structure of GPR34-Gi complex
Method: single particle / : Yong XH, Zhao C, Yan W, Shao ZH

EMDB-35347: 
Structure of R2 with 3'UTR and DNA in binding state
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

EMDB-35348: 
Structure of R2 with 3'UTR and DNA in unwinding state
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

EMDB-35349: 
Structure of R2 with 5'ORF
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

EMDB-35350: 
Structure of R2 with 5'ORF and 3'UTR
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

PDB-8ibw: 
Structure of R2 with 3'UTR and DNA in binding state
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

PDB-8ibx: 
Structure of R2 with 3'UTR and DNA in unwinding state
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

PDB-8ibz: 
Structure of R2 with 5'ORF and 3'UTR
Method: single particle / : Deng P, Tan S, Wang J, Liu JJ

EMDB-36300: 
Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36312: 
Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36317: 
Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36318: 
Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36490: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36491: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36492: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36493: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36494: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36495: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36496: 
Cryo-EM map of human receptor R2
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH

EMDB-36497: 
Cryo-EM map of human Gi heterotrimer
Method: single particle / : Zhao C, Tian XW, Liu Y, Cheng L, Yan W, Shao ZH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
